- Description:
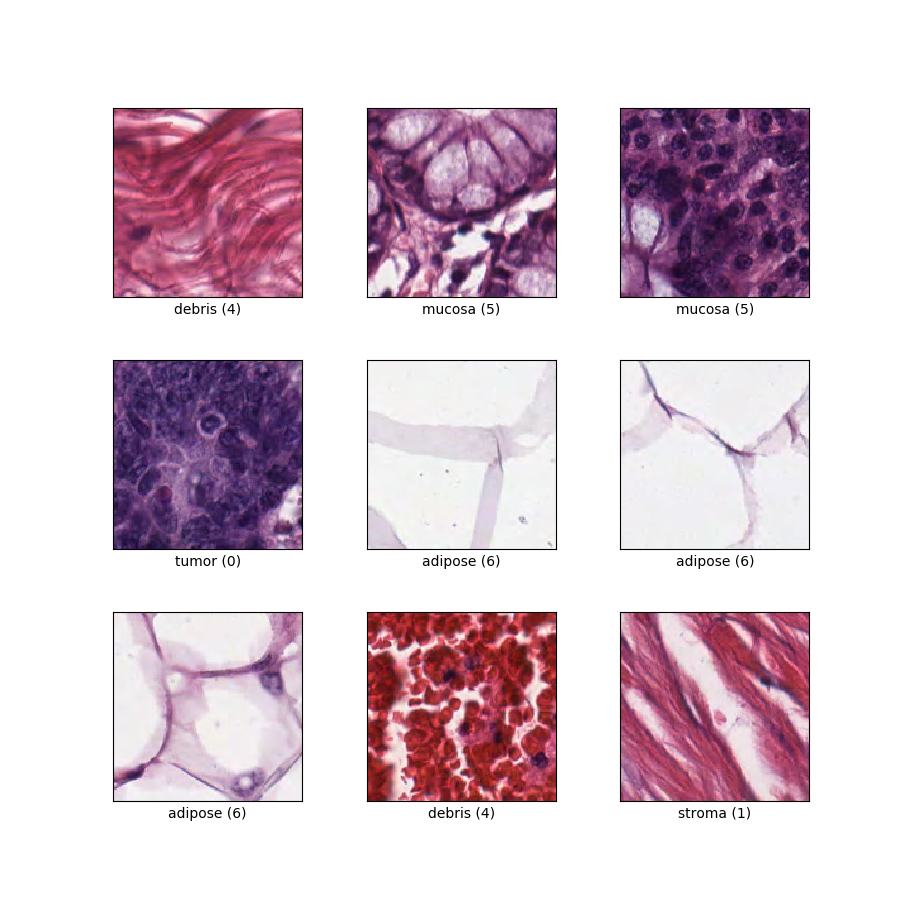
Classification of textures in colorectal cancer histology. Each example is a 150 x 150 x 3 RGB image of one of 8 classes.
Source code:
tfds.image_classification.ColorectalHistologyVersions:
2.0.0(default): New split API (https://tensorflow.org/datasets/splits)
Download size:
246.14 MiBDataset size:
179.23 MiBAuto-cached (documentation): Only when
shuffle_files=False(train)Splits:
| Split | Examples |
|---|---|
'train' |
5,000 |
- Feature structure:
FeaturesDict({
'filename': Text(shape=(), dtype=string),
'image': Image(shape=(150, 150, 3), dtype=uint8),
'label': ClassLabel(shape=(), dtype=int64, num_classes=8),
})
- Feature documentation:
| Feature | Class | Shape | Dtype | Description |
|---|---|---|---|---|
| FeaturesDict | ||||
| filename | Text | string | ||
| image | Image | (150, 150, 3) | uint8 | |
| label | ClassLabel | int64 | Eight classes: 0: 'tumour epithelium', 1: 'simple stroma', 2: 'complex stroma' (stroma that contains single tumour cells and/or single immune cells), 3: 'immune cell conglomerates', 4: 'debris and mucus', 5: 'mucosal glands', 6: 'adipose tissue', and 7: 'background'. |
Supervised keys (See
as_superviseddoc):('image', 'label')Figure (tfds.show_examples):
- Examples (tfds.as_dataframe):
- Citation:
@article{kather2016multi,
title={Multi-class texture analysis in colorectal cancer histology},
author={Kather, Jakob Nikolas and Weis, Cleo-Aron and Bianconi, Francesco and Melchers, Susanne M and Schad, Lothar R and Gaiser, Timo and Marx, Alexander and Z{"o}llner, Frank Gerrit},
journal={Scientific reports},
volume={6},
pages={27988},
year={2016},
publisher={Nature Publishing Group}
}
